 |
 |
||||
| BrainSqueezer | |||||
Display and Coregister multiple 3D images |
|||||
 |
 |
||||
| BrainSqueezer | |||||
Display and Coregister multiple 3D images |
|||||
Coregistration Features:
There are many approaches to coregistration of one image volume into the spatial coordinates of another. At the Keck Lab, we use Roger Wood's AIR package (UCLA), AFNI (Bob Cox), the coregistration suite within SPM99 (Wellcome Group, London), FSL (Oxford), and MPITool (Advanced Tomo Vision GmbH). Most of these are automated in the sense that they attempt to find an optimal set of parameters describing how one volume will best fit onto another one. Suffice it to say that these methods work really well for normal brains and good-quality scans, but increasingly less well as we stray farther from ideal conditions. And it wouldn't really be research if everything was perfect, now, would it?
BrainSqueezer fills the niche of coregistering difficult scans, where you can see how they should be aligned but automated methods just can't get it right. BrainSqueezer also provides a number of tools for checking the accuracy of a coregistration performed by another package. This is important, since most automated methods have some level of innaccuracy. Of course, as long as you have to check the accuracy, you might as well just use BrainSqueezer to do the coregistration in the first place...
 |
Standard Display:
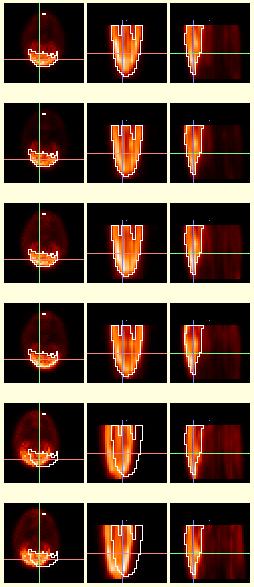
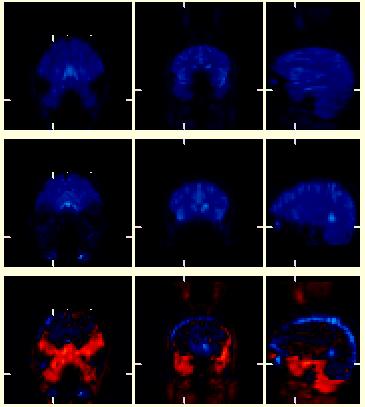
To the left is a series of image volumes showing [18F]-FDG concentration in six (6) different monkeys, taken from a larger set of 20 PET scans. All 20 scans were read and displayed simultaneously by BrainSqueezer, but only a subset is shown here for conciseness. The left column shows the coronal slices (the top of the brain is at the bottom of the picture in this particular view), the middle column shows the axial slices (the anterior portion of the brain is at the bottom of the screen), and the right column shows the sagittal slices (anterior at bottom). The field of view of the PET scanner covered the anterior half of the brain. The pixel dimensions of the images in this example are not to scale. This is the standard display within BrainSqueezer, showing 3 orthogonal views. The crosshairs are color-coded so that the red line denotes the X-axis (right-left), green is the Y-axis, and blue is the Z-axis. The value at the center of the crosshairs for each volume is displayed to the right of the right-hand column (not shown here).The contour (in white) around the brain is taken from the top reference volume and applied to all of the other volumes. The monkeys were anesthetized and a head-holder was used for consistent location within the PET scanner. Particular emphasis was placed on putting the amygdala in the same axial plane within the PET scanner. However, the bottom two scans are not perfectly aligned with the top scan. Using this display format it is easy to see the type and extent of misalignment, which consists primarily of a shift to the left. The automated coregistration packages we tried could not align these images to our satisfaction.What makes them challenging is that only half of the brain is present in these data, and the false edge in the middle of the brain is one of the most dominant features. However, using BrainSqueezer we were able to rapidly and accurately align these problematic images. |
| Image Math:
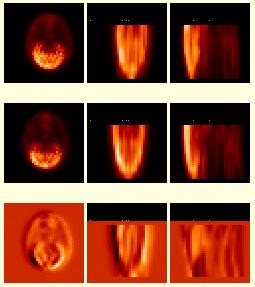
BrainSqueezer can create a "Math Image" using two image volumes. The picture to the right shows a "reference" image volume on the top, an "object" volume in the middle, and on the bottom is the result of subtracting the object volume from the reference volume. The bottom row always shows the "Math" image, and can change depending on the mathematical operation you select. Subtraction or ratio images can be particularly helpful to distinguish subtle misalignments that may not be apparent with a contour overlay. For example, there is a pronounced shift to the left of the object volume, evident from the lighter band on the right-hand side of the coronal (left) subtraction image. Other Math Image operations include addition, multiplication, and division. Multiplication is useful to "mask off" a portion of one image with another. Division is useful to create a ratio-image, which is particularly handy if you want to compare two image volumes which differ by a large offset. There is |
 |
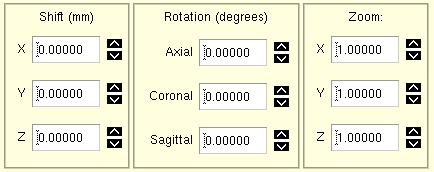 |
Perhaps the most useful thing about BrainSqueezer (and the source of its name), is its ability to TAKE ACTION if you see that you do not like how one image is aligned to another. The control panel for moving one volume relative to another is shown to the left. There are two modes: a fast slice-wise 2D method, and a completely IDL-run 3D method that uses trilinear interpolation. |
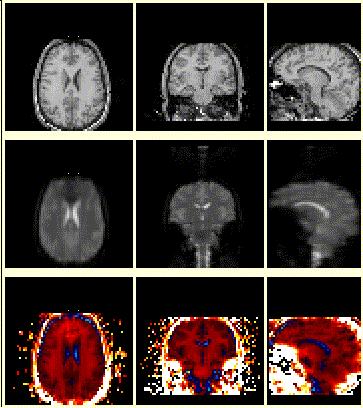
| The images to the right show a series of EPI (fMRI-BOLD) image volumes. The top images were acquired as a series of coronal planes (left:coronal, middle:axial, right:sagittal). The middle volume is a similar EPI scan of the same person but the planes are "tilted" or acquired at an oblique angle relative to the top volume. The bottom images show a subtraction image of (top - middle), with positive differences coded in various shades of red, and negative differences coded in shades of blue. These image volumes started life with completely different dimensions, and demonstrate the ability of BrainSqueezer to read and display such files. Displayed in this manner, it is easy to see how one volume needs to be moved to become aligned with the other. In addition, the so-called dropout artifact, where signal is lost due to surrounding tissue inhomogeneity, can be easily seen in the areas colored red. Since these two volumes have not yet been coregistered the red region is also due to a simple misalignment, but subsequent analysis shows that most of the difference highlighted by the red color is due to differences in dropout artifact. |
 |
 |
And, of course, a coregistration program wouldn't be worth the magnetic dipoles it is written on if it couldn't perform inter-modality alignment. It was a major breakthrough in the early '90s when automated programs were developed to do this. The main issue is that the image values and the distribution of these values can be vastly different between modalities. BrainSqueezer does not care if the images are intra-, inter-, or Martian modality. There is nothing within the program that makes any kind of judgement about the alignment of the images. It is completely up to you. BrainSqueezer just gives you the tools to shove the images around and to judge how a good a job you did. And it doesn't take as long as you might think... |