Adaptive Cuts for Extracting Specific White Matter Tracts
Authors: Nagesh
Adluru, Vikas Singh, Andrew L. Alexander
Acknowledgement: The work is supported in part by the following grants:
NSF IIS 1116584, NIH R21 AG034315, NIH P50-MH84051, NIH R01 MH08026, NIH P30 HD003352 and the Henry M. Jackson Foundation.
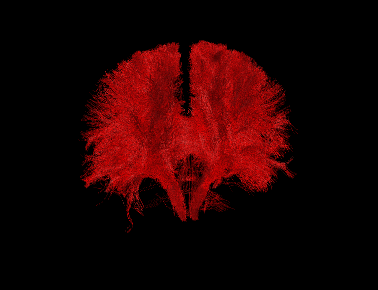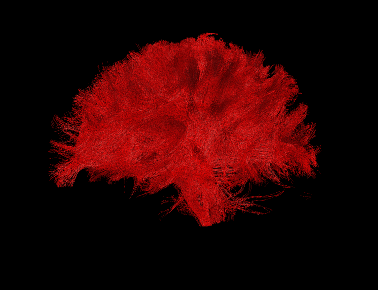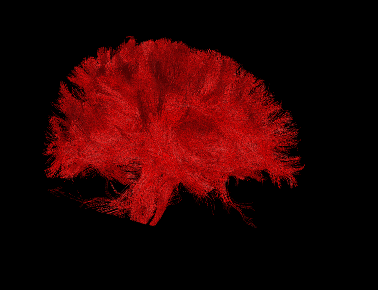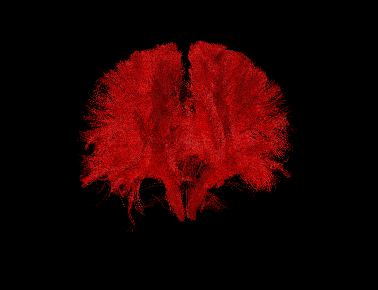
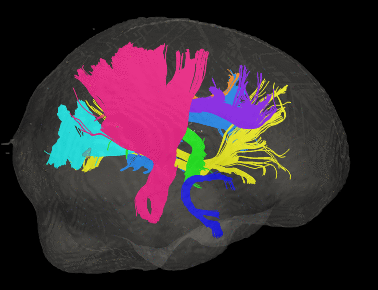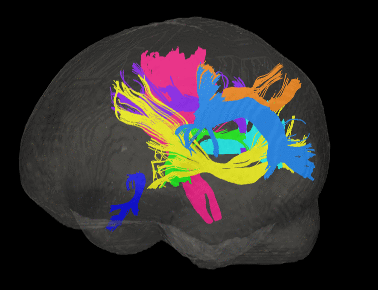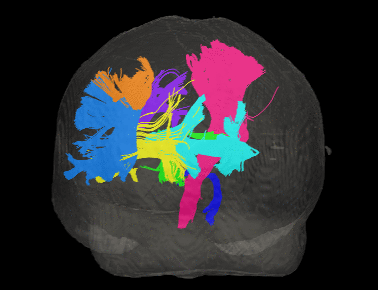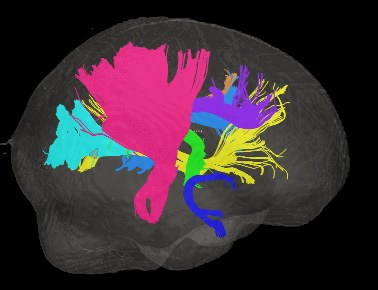
Basic overview:
from whole brain tractography has numerous applications
in studying individual differences in white matter. Typically
specific tracts are extracted manually, following replicable
protocols which can be prohibitively expensive for large scale
studies. A tract clustering framework is a suitable computational
framework but from a neuroanatomical point of view,
one of the key challenges is that it is very hard to design a
universal similarity function for different types of white matter
tracts (e.g., projection, association, commissural tracts). In
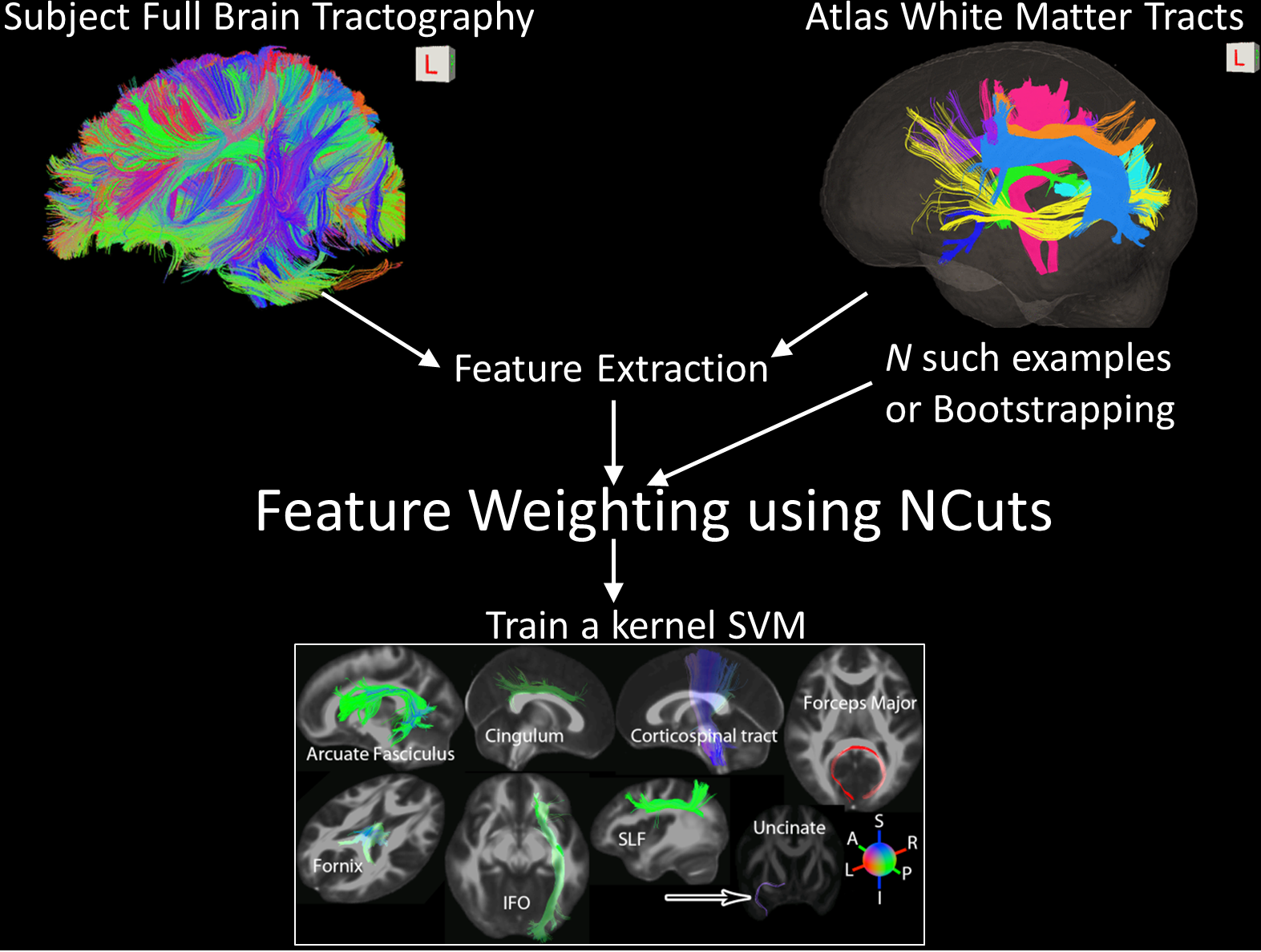
this paper, we propose an adaptive cuts framework in which,
using normalized cuts motivated objective function, we adaptively
learn tract-tract similarity for each specific tract class
using atlas based training data. Using the learnt similarity
function we train an ensemble of binary support vector machines
to extract specific tracts from unlabeled whole-brain
tractography sets.
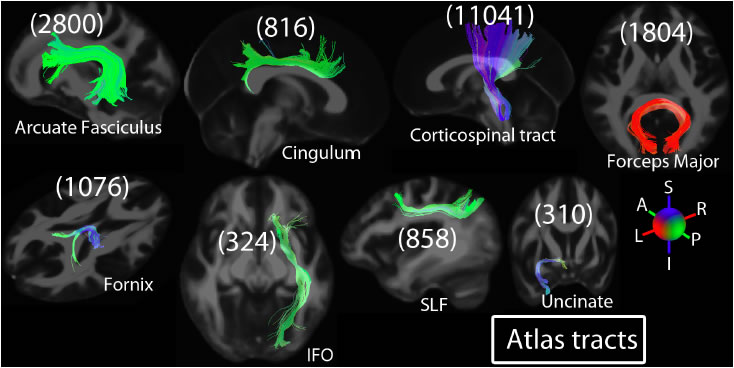
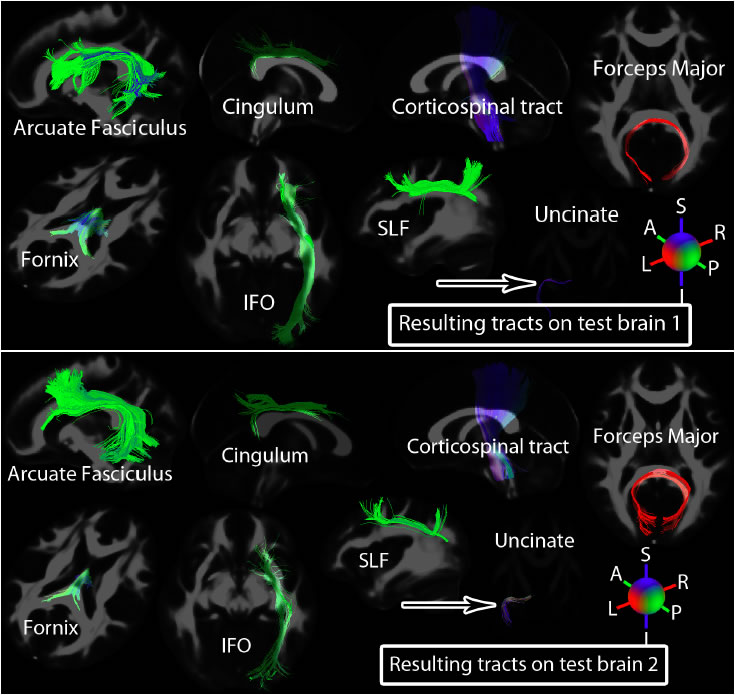
Sample Results:
Code available upon request (nagesh.avr@gmail.com).
Snapshot of sample usage of the code:
PIPELINE
%------------------------------------------------------------------------------------
%Training component (on the atlas).
%Feature extraction.
script_compute_tract_features
%Generating training data for feature weighting.
script_bootstrap_training_v4
script_visualize_bootstrapping_v4
%Learning the feature weighting.
main_call_file_tracts_v5 -> getUVTracts_v3, feature_weigh_convex_v2
script_visualize_K_trains (before learning alphas)
script_visualize_alphas
main_call_file_tracts_v5_debugging (for optimization checks)
%SVM.
script_ensemble_SVM_train_prep (train-train weighted kernels)
script_ensemble_SVM_train -> get_loo_vals
%------------------------------------------------------------------------------------
%Testing component (on a new subject).
%Feature extraction.
script_compute_tract_features
%Applying the learnt SVM on to the test set.
script_ensemble_SVM_classify_prep (test-train weighted kernels)
script_ensemble_SVM_classify
%Visualizing results.
scrtip_write_output_trks -> write_output_bundles.m
%For Evaluating results using volumetric overlaps.
for i in `ls brain1_scan1_*.trk`;do prefix=`echo $i | awk 'BEGIN{FS="."}{print $1}'`; echo $prefix; trackvis_to_camino -i $i -o ${prefix}.Bfloat;cat ${prefix}.Bfloat | tractstatimage -outputroot ${prefix} -scalarfile FMRIB58_FA_1mm.nii.gz -countintersect -tractstat sum;fslmaths ${prefix}.hdr -bin ${prefix};rm -f ${prefix}.hdr ${prefix}.img;done