Introduction
Based on the Weighted Fourier Series (WFS)
framework originally developed for brain cortical
surfaces [1] and amygdala surfaces [2], white
fiber tracks are parameterized using the cosine
basis functions. This is a general framework for
representing, registering and smoothing anatomical
objects in a unified Hilbert space formulation.
The cosine series representation for 3D curves is
introduced in [3] and [5]. The implementation is
based on MATLAB 7.5 and a Mac-OSX. If you are using the codes or
data below, please reference [3] or [5]
Loading tract data to MATLAB
Using the second order Runge-Kutta streamline algorithm with tensor deflection (TEND) [4], we obtained the whole brain tracts (001_spd_tracts.Bfloat.zip) for a single subject. You can use CAMINO package to obtain tracts. The complete MATLAB package excluding the tract data is zipped into cosine-representation.zip. After unzipping, load the tract. The variable SL consists of 10000 tracts and 1000-th tract can be displayed:SL=get_streamlines('001_spd_tracts.Bfloat',[1.5 1.75 2.25]);
tract=SL{1000}';
figure; plot3(tract(1,:),tract(2,:),tract(3,:),'.b')
Cosine series representation
The
parameterization of the tract is done by[arc_length para]=parameterize_arclength(tract);
Note that this is not the arclengh paramterization. Instead we paramterize the arclength as a unit interval. The i-th control point in the tract tract(:,i)is mapped to para(i), where para is a number between 0 and 1:
figure;
subplot(3,1,1); plot(para, tract(1,:));
subplot(3,1,2); plot(para, tract(2,:));
subplot(3,1,3); plot(para, tract(3,:));
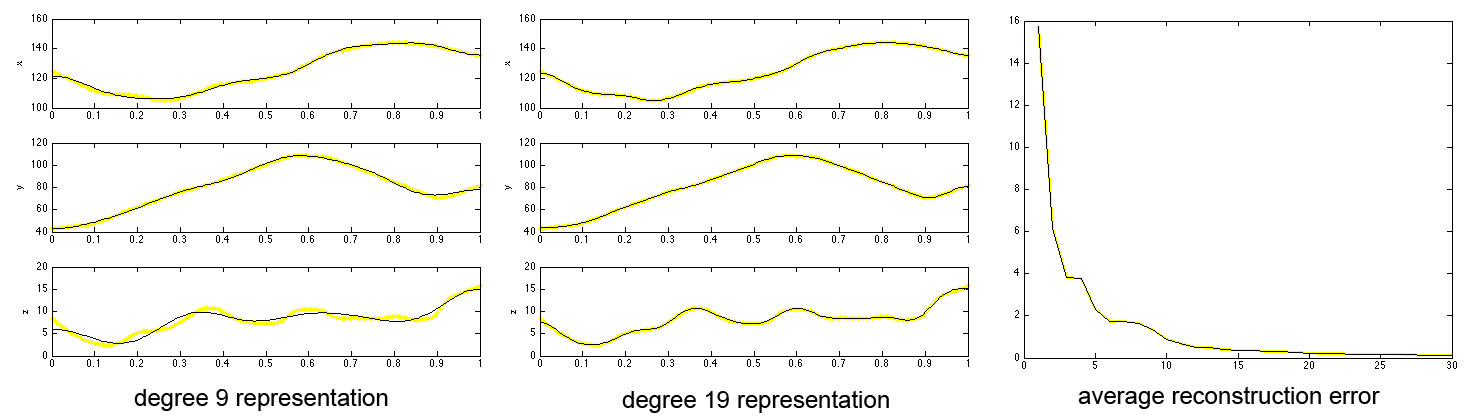
Figure 1. Plots of x,y,z coordinates of the cosine series representation. Yellow lines are the original tract coordinates (tract) and black lines (wfs) are the reconstruction. The average reconstruction error over degree is given in terms of mean sum of squared errors over whole tract. See [3] for detail.
The 19-th degree cosine series representation is given by
[wfs beta]=WFS_tracts(tract,para,19);
figure; plot3(tract(1,:),tract(2,:),tract(3,:),'.y') %original data
hold on; plot3(wfs(:,1),wfs(:,2),wfs(:,3),'-k') %reconstruction
beta is the estimated Fourier coefficients and wfs is the coordinates of the representation.
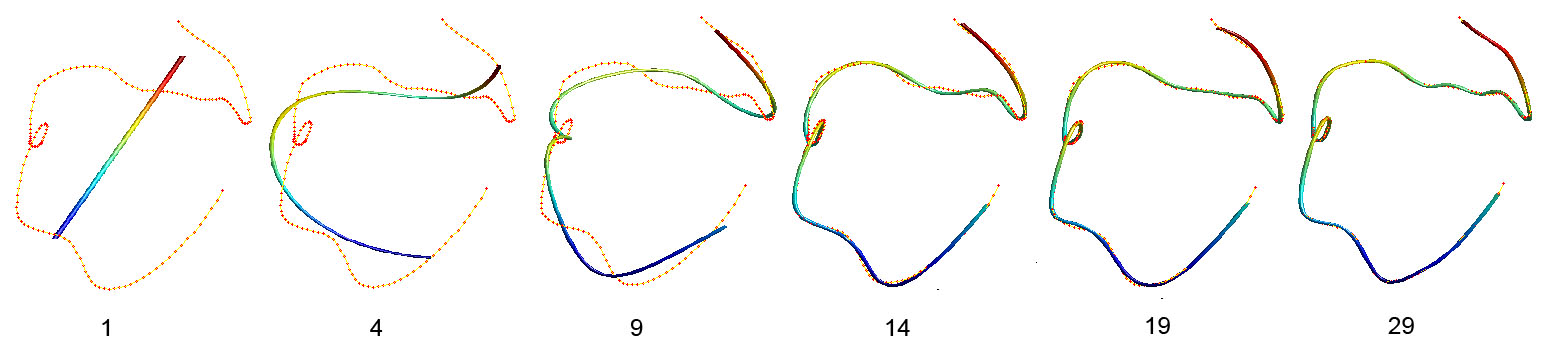
Figure 2. The cosine series representation at various degrees. The orange dots are variable tract while the colored lines are variable wfs.
Averaging curves
The cosine series representation can be used to
average the collection of curves by simply
averaging the coefficients. Consider another tract
generated by adding noise:
tract2 = tract +
normrnd(0,0.5,3,491);
hold on;
plot3(tract2(1,:),tract2(2,:),tract2(3,:),'.y')
[arc_length2 para2]=parameterize_arclength(tract2);
[wfs2 beta2]=WFS_tracts(tract2,para2,19);
hold on;
plot3(wfs2(:,1),wfs2(:,2),wfs2(:,3),'-k')
%reconstruction
It is
nonsense to simply average tract
and tract2 by averaging their
coordinates. Instead, we simply average the
representation. Using the average
coefficients, we can reconstruct the cosine
representation using WFS_resample.
beta_a= (beta + beta2)/2;
para_a = [1:200]/200;
wfs_a=WFS_resample(beta_a,19, para_a);
hold on;
plot3(wfs_a(:,1),wfs_a(:,2),wfs_a(:,3),'-r')
Simulating 3D curves
This is an example given in
[3]. Taking this as the underlying signal, noises
are added nonlinaerly to obtain the simulation
results.
t=0:0.1:10
tract=[t.*sin(t); t.*cos(t); t];
figure;
plot3(tract(1,:),tract(2,:),tract(3,:),'.b')
%simulation
[arc_length para]=parameterize_arclength(tract);
[wfs beta]=WFS_tracts(tract,para,19);
hold on; plot3(wfs(:,1),wfs(:,2),wfs(:,3),'r')
%reconstruction
Streamtube representation
Figure 7 in paper [3] has a very interesting
visualization technique called the streamtube
representation. This is done using the built-in
function called streamtube in MATLAB.
figure;
figure_streamtube(tract',
0.5, [1 .3 .3]);
camlight
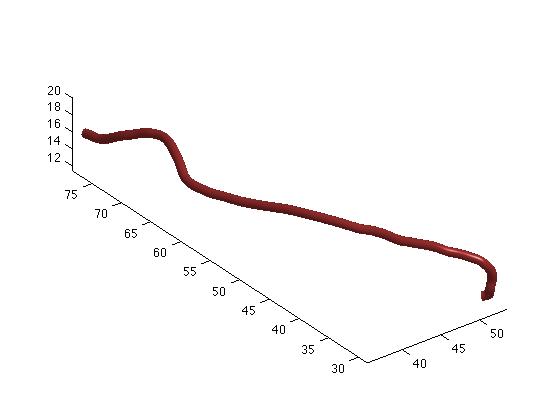
Figure 3. The streamtube representation of a tract.
This generates Figure 3. If you use this function multiple times while holding the figure using the hold on command, you can generate some amazing 3D tract visualization (Figure 4).
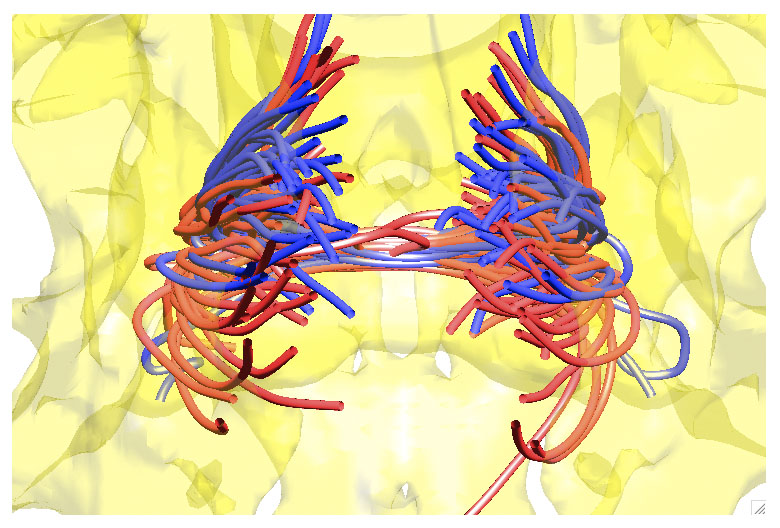
Figure 4. The streamtube representation of a bundle of tracts for two groups (red= autism, blue = controls). See paper [3] for details.
Weighted Fourier series (WFS) representation
The terms in the cosine series representation can
be exponentially weighted in such a way that the
expansion is the solution of heat diffusion or
equivalently heat kernel smoothing. For details,
see [1].
We will demonstrate this using EEG time series published in [6]. The representation
can be used to smooth out noisy EEG (Figure 5).
load EEG.mat
x=[1:3500]/3500;
k=2000;
sigma=0.0001;
[wfs beta]=WFS_COSINE(EEG(1,:)',x',k,sigma);
figure; plot(EEG(1,:), '.k');
hold on; plot(wfs, 'r', 'linewidth',1)

Figure 5. Weighted Fourier series representation of EEG, which is equivalent to heat diffusion. Left is with bandwidth 0.00001 and right is with bandwidth 0.0001.
References
[1] Chung, M.K., Dalton, K.M., Shen, L., L., Evans, A.C., Davidson, R.J. 2007. Weighted Fourier series representation and its application to quantifying the amount of gray matter. IEEE Transactions on Medical Imaging 26:566-581.[2] Chung, M.K. Nacewicz, B.M., Wang, S., Dalton, K.M., Pollak, S., Davidson, R.J. 2008. Amydala surface modeling with weighted spherical harmonics. 4th International Workshop on Medical Imaging and Augmented Reality (MIAR). Lecture Notes in Computer Sciences (LNCS) 5128:177-184.
[3] Chung, M.K. Adluru, N., Lee, J.E., Lazar, M., Lainhart, J.E., Alexander, A.L. 2010. Cosine series representation of 3D curves and its application to white matter fiber bundles in diffusion tensor imaging. Statistics and Its Interface 3:69-80.
[4] Lazar, M., Alexander, A.L. 2005. Bootstrap white matter tractography (BOOT-TRAC). NeuroImage 24:524-532.
[5] Chung, M.K. 2013. Statistical and Computational Methods in Brain Image Analysis. CRC Press. DATA/MATLAB. The detail is given in Chapter 10.
[6] Wang, Y., Chung, M., Bachhuber, D.R.W., Schaefer, S.M., van Reekum, C.M., Davidson, R.J. 2015 LARS network filtration in the study of EEG brain connectivity, IEEE 12th International Symposium on Biomedical Imaging (ISBI) 30-33